
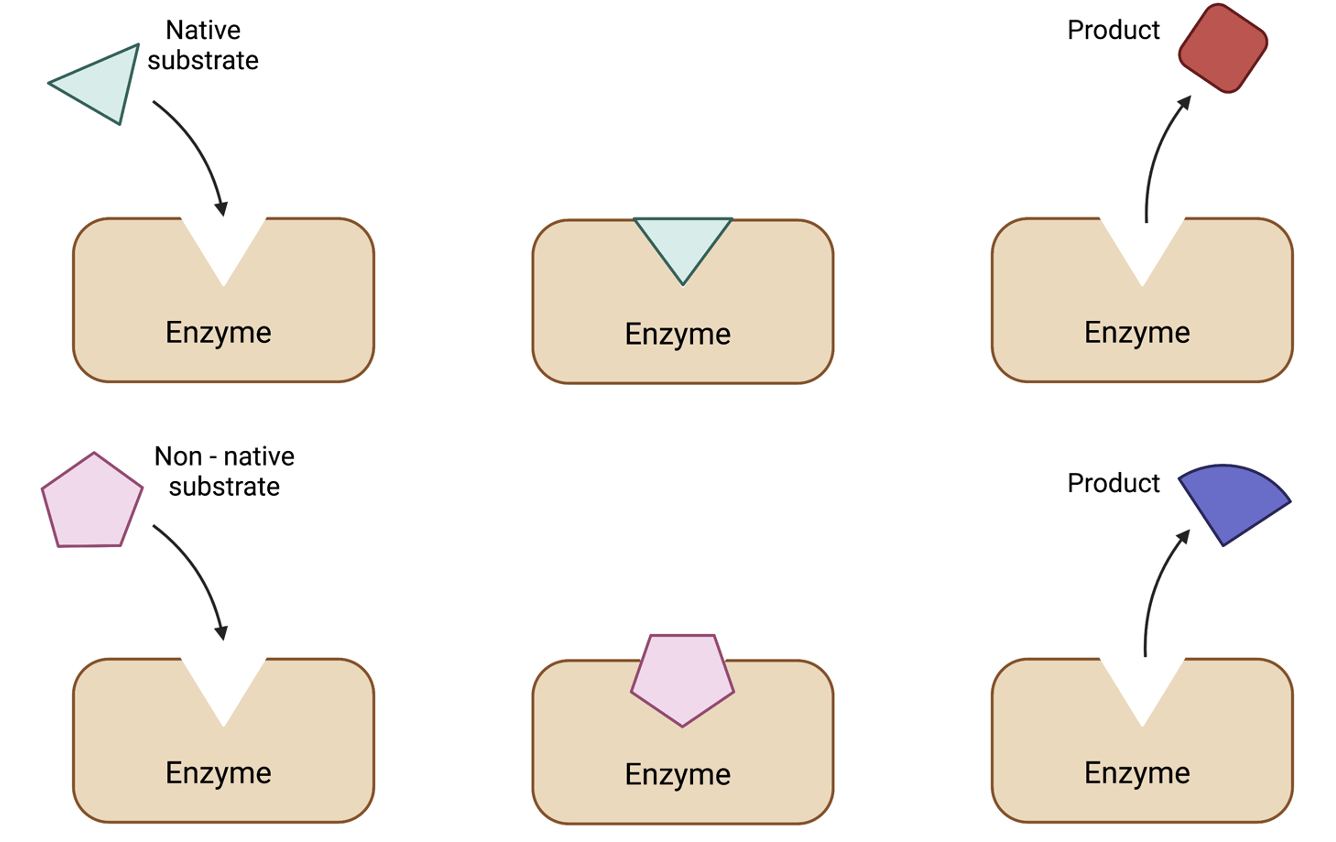
1. Catalysis - Metabolic Engineering
Cells are capable of exquisite catalysis. By consuming a wide variety of raw materials, cells are able to synthesize alkanes, sophisticated small molecules, and high information content polymers (DNA, RNA, protein) that fold into complex three-dimensional structures. Broadly, our efforts focus on introducing heterologous enzymes to microbes in order to perform the chemical reactions we desire as well as developing control strategies to balance native metabolic pathways with foreign enzymes to maximize product yield.
We are very interested in using the tools of synthetic biology to develop metabolic engineering strategies that are robust to cell biology and adaptable to a variety of engineering goals. For example, we are currently looking at how cellular stress responses may be sensed and controlled in order to best improve product yields (Rashmi). Additionally, another project focuses on harnessing the enzyme promiscuity of classes of enzymes to create interesting and valuable new compounds with complex chemistry (Bradley, Tracey).
We use these capabilities to produce small molecule drugs, protein therapeutics, and fuels from very cheap, renewable raw materials. Current projects focus on the use of lignin derivatives and engineering the organisms capable of utilizing these cheap and renewable feedstocks (Bradley, Erika), as well as nitrogen-rich animal waste streams (Amin).
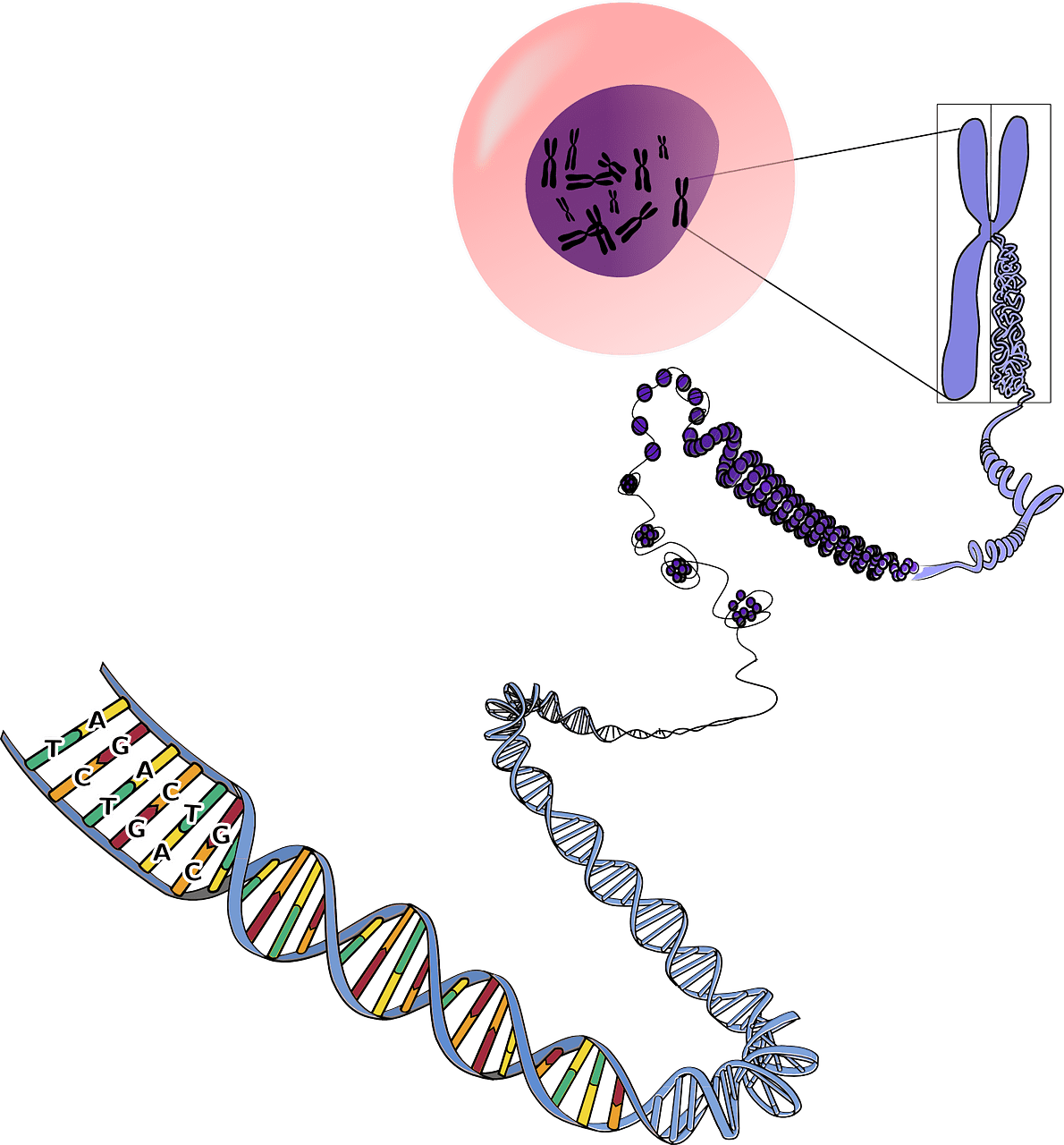
2. Sensing - Signaling Engineering
Microbes must accurately sense the environment around it to protect itself from harm and capitalize on nutrient opportunities. Microbial sensing is highly effective because it is specific and sensitive and produce a desired cellular response in a timely manner.
We use these capabilities to engineer cells that can detect new environmental cues. For example, current projects with DNA polymerase-based biosensors under development in the lab have the potential to improve the scale and resolution of neural activity measurements in vivo (Alec, Namita, and Marija), and also to better understand the mechanisms of these enzymes through modeling (Jon). We also focus on the detection of biomarkers that currently do not have effective low-cost diagnostics for potential use in resource-poor settings (Cat).
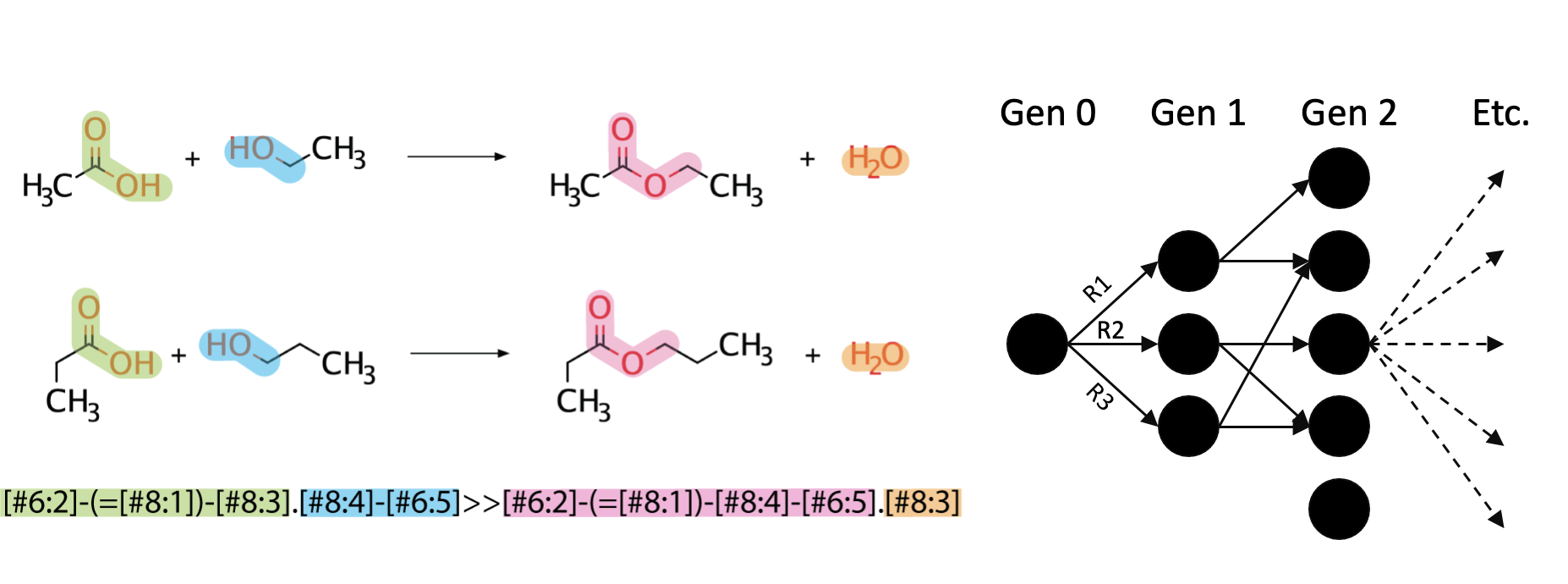
3. Uniting computational predictions with experimental applications
To accomplish these aims we bring together expertise ranging from in silico modeling to genetic engineering and experimental validation. Computational resources, designed by and developed in conjunction with the Broadbelt lab, are currently being used to direct the design of novel biosynthetic pathways. These projects include understanding the fundamental rules of biochemistry and mapping those to protein features (Joseph), as well as designing pathways which will allow for the biocatalytic upgrading of precursors obtained from traiditional chemistry (Kevin).
Another area of interest in our computational research is using machine learning to understand how enzyme promiscuity can be predicted from substrate features. A current project in this area is aiming to predict carboligase activity from multiple substrates using support vector machines (Tracey).
Lastly, in silico simulations such as Flux Balance Analysis and Ensemble Modeling can also tell us more about how perturbations of the metabolome will affect a cell and the efficiency of proposed synthetic routes. Some current projects include flux balance analysis of non-model organisms on renewable substrates (Jon), as well as ODE modeling of metabolism in cell-free systems to facilitate rapid metabolic engineering pipelines (Jake).
